Change Durations Spm Mat

Stats maxmem maximum amount of data processed at a time in bytes spm spmid version of spm used spm swd directory for spm mat and img files.
Change durations spm mat. Mask img hdr analysis mask image image of zeroes ones indicating which voxels were included in the analysis. However a good approach is to use the gui to set up an example analysis then fetch the view the resulting m code there is an option in the gui menu. Saving spm regressor file d 20161020ys analyse functional art regression outliers waus20161020ys 0005 00001 000001 01 mat and d 20161020ys analyse functional art regression outliers and movement waus20161020ys 0005 00001 000001 01 mat. Note that some features in bspmview will be automatically disabled if you load in a statistical image that was generated outside of spm for instance in fsl or if bspmview is unable to locate the model output files e g spm mat resms nii associated with the provided statistical image.
Reads img hdr nii and nii gz. Stats maxres maximum number of residual images for smoothness estimation spm. Specify effects of real interest name. This mask image is the intersection of the explicit implict and threshold masks specified in the xm argument.
5 12 2017 1 comment if you are using spm for fmri data analysis and entering onsets and durations for each condition in the first level manually then you should consider using the multiple conditions option and input a mat file that has all the onsets and durations. Generating onset and duration mat file for spm for fmri analysis. I ve figured out matlab doesn t seem to allow changing the values within the cell it gave me strange errors indicating that the entire file no longer existed when i tried to change them. You can browse through its fields and we then use the spm desrep function to display the design matrix and other 1st level statistical information this does the same as when you use the spm12 gui review option and select the spm mat file.
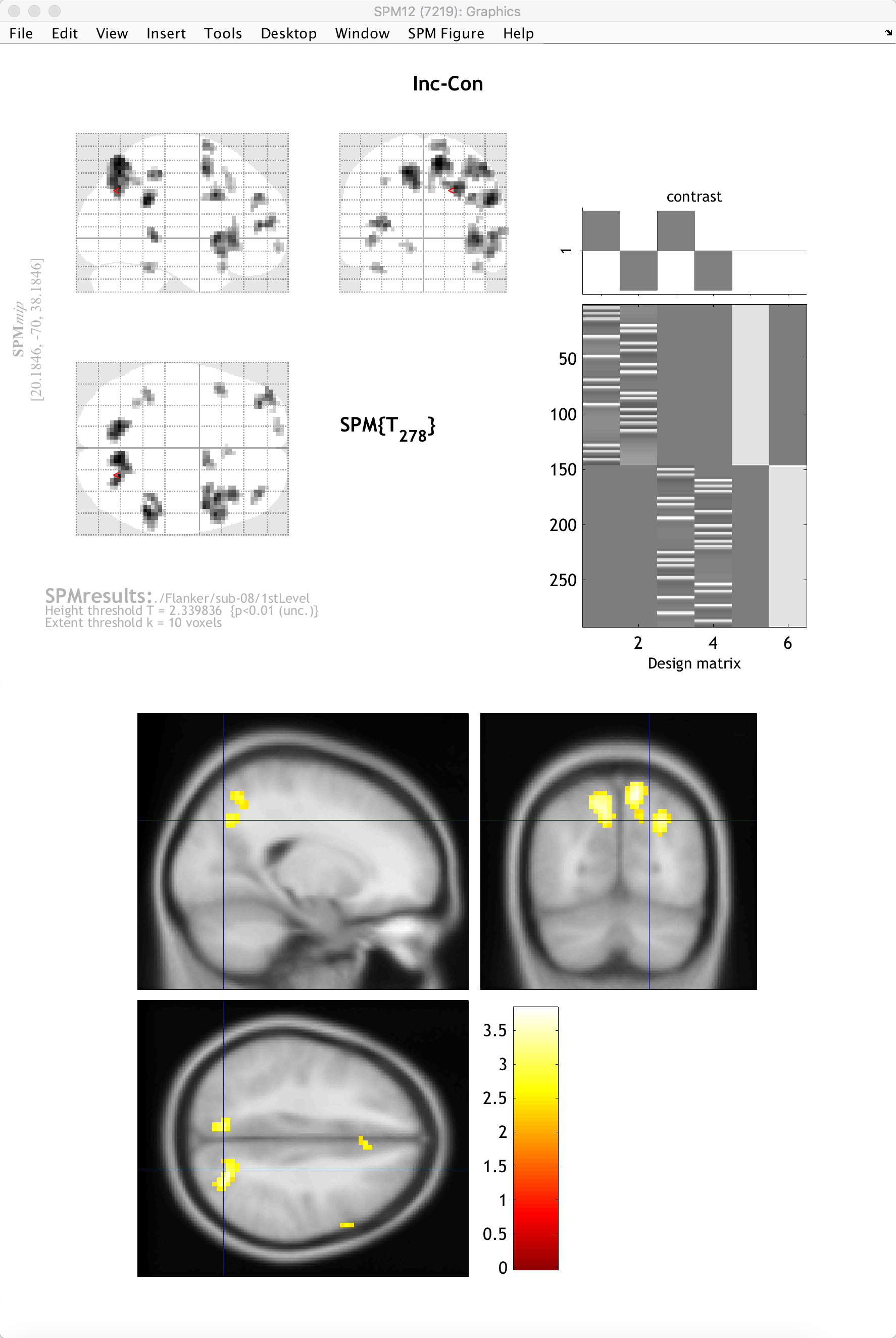
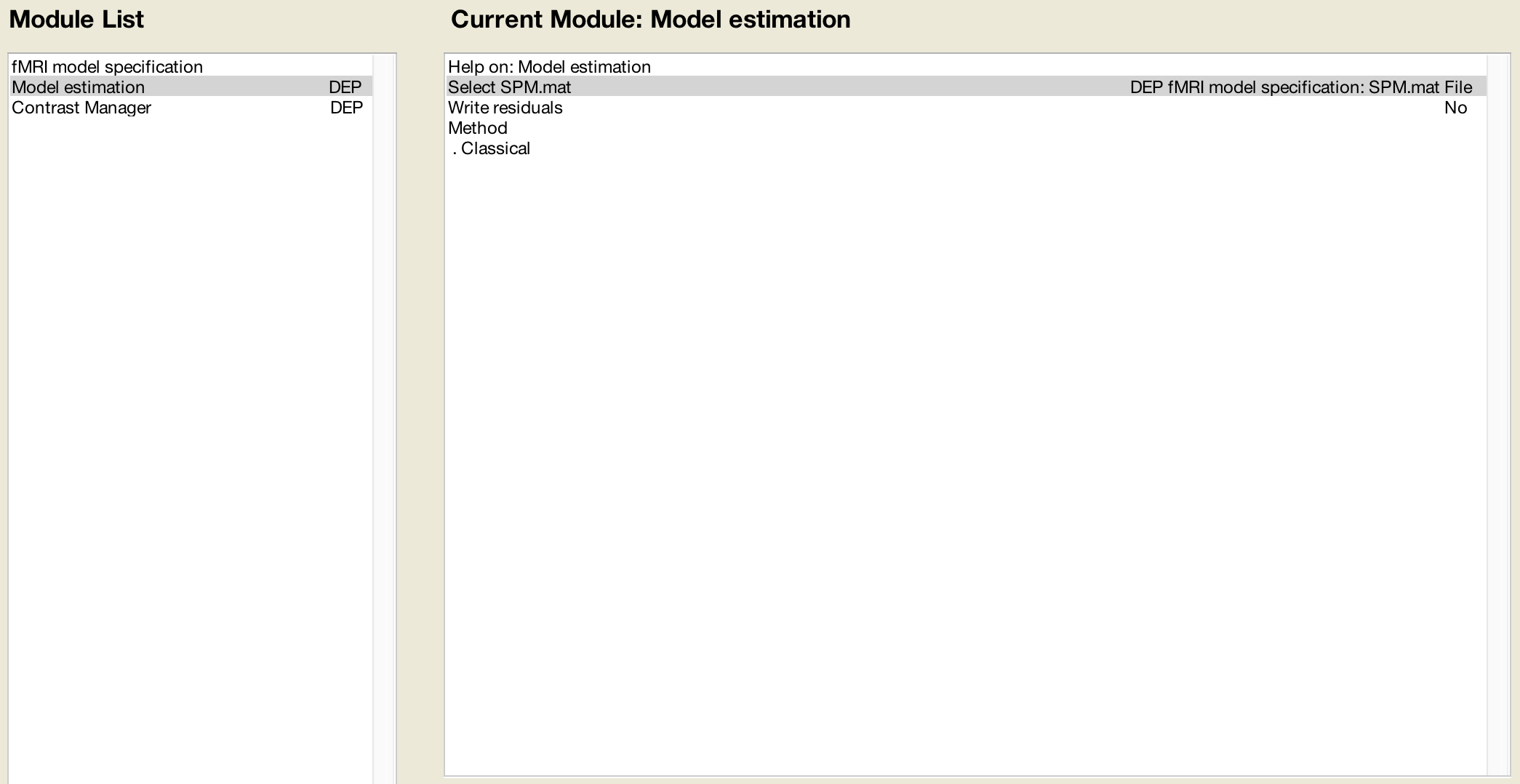
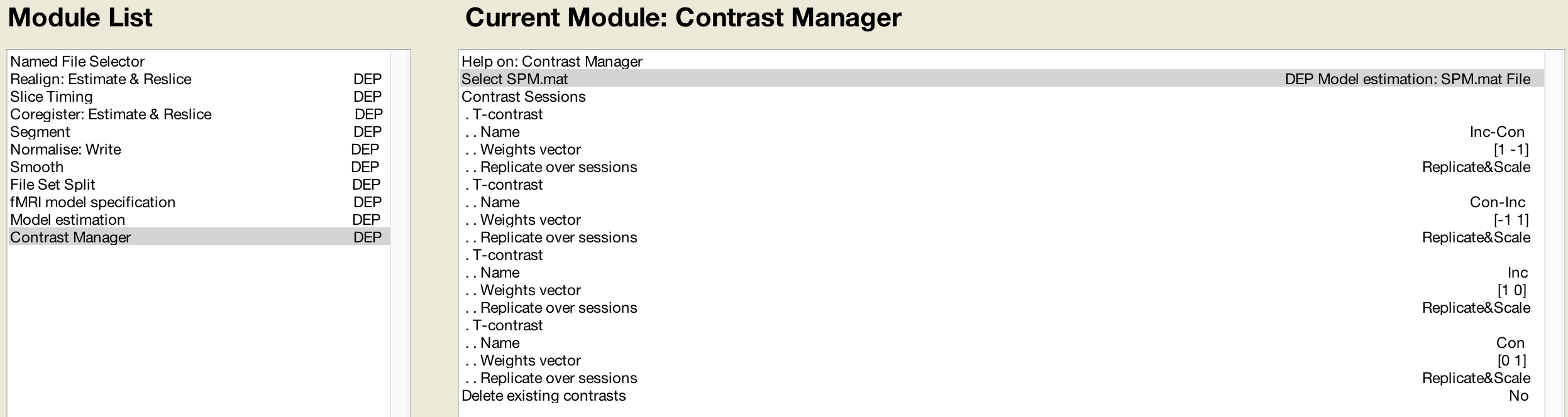
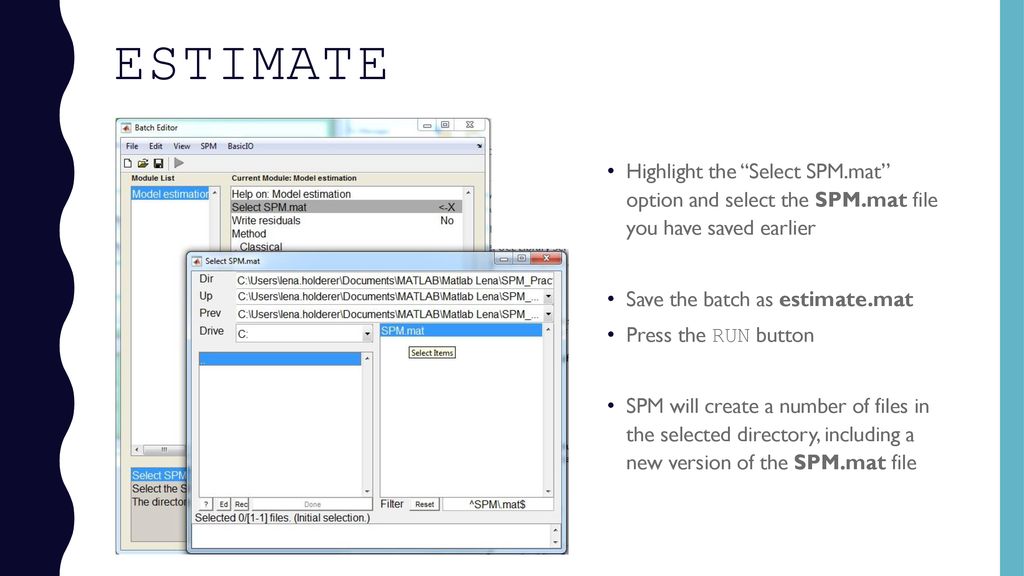
You can use it to inspect the design. After spm mat estimation the following images are written to file. Press results select the spm mat file press f contrast in the contrast manager and select define new contrast. This spm structure is loaded into the matlab workspace i e.
Spm xbf name name of basis function spm xbf length length in seconds of basis. I am trying to change spm preprocessing parameters stored in a mat file. If you change the location of your data and or the spm installation the full paths contained in batch files and spm mat will be broken. Documentation on batch processing in spm is less complete than one would like.